Welcome
Contents
Welcome#
We type code in cells and then run them.#
2 + 4
6
2 ** 4
16
days_in_week = 7
hours_in_week = days_in_week * 24
hours_in_week
168
We use libraries other people wrote#
# Some code to set up our notebook for data science!
from datascience import *
from cs104 import *
import numpy as np
%matplotlib inline
We write code to manipulate and analyze data#

hopkins_trees = Table.read_table("data/hopkins-trees.csv")
hopkins_trees.show(10)
| plot | genus | species | common name | count |
|---|---|---|---|---|
| p00-1 | Acer | pensylvanicum | Maple, striped | 28 |
| p00-1 | Acer | rubrum | Maple, red | 8 |
| p00-1 | Acer | saccharum | Maple, sugar | 12 |
| p00-1 | Amelanchier | canadensis | Shadbush | 17 |
| p00-1 | Betula | alleghaniensis | Birch, yellow | 7 |
| p00-1 | Betula | papyrifera | Birch, paper | 5 |
| p00-1 | Fagus | grandifolia | Beech, American | 142 |
| p00-1 | Ostrya | virginiana | Hophornbeam | 7 |
| p00-1 | Prunus | serotina | Cherry, black | 11 |
| p00-1 | Quercus | rubra | Oak, red | 6 |
... (3783 rows omitted)
hopkins_trees = hopkins_trees.drop("genus", "species")
hopkins_trees
| plot | common name | count |
|---|---|---|
| p00-1 | Maple, striped | 28 |
| p00-1 | Maple, red | 8 |
| p00-1 | Maple, sugar | 12 |
| p00-1 | Shadbush | 17 |
| p00-1 | Birch, yellow | 7 |
| p00-1 | Birch, paper | 5 |
| p00-1 | Beech, American | 142 |
| p00-1 | Hophornbeam | 7 |
| p00-1 | Cherry, black | 11 |
| p00-1 | Oak, red | 6 |
... (3783 rows omitted)
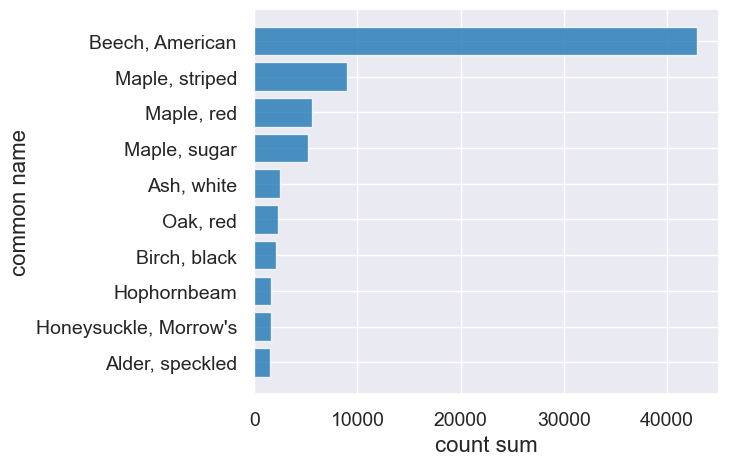
How many of each species?#
hopkins_trees.drop("plot").group("common name", sum).sort("count sum", descending=True)
| common name | count sum |
|---|---|
| Beech, American | 42922 |
| Maple, striped | 8939 |
| Maple, red | 5564 |
| Maple, sugar | 5193 |
| Ash, white | 2523 |
| Oak, red | 2283 |
| Birch, black | 2144 |
| Hophornbeam | 1613 |
| Honeysuckle, Morrow's | 1608 |
| Alder, speckled | 1564 |
... (67 rows omitted)
tree_counts = hopkins_trees.drop("plot").group("common name", sum)
tree_counts = tree_counts.sort("count sum", descending=True)
tree_counts.take(np.arange(0,10)).barh("common name")
Where are all the red maples?#
red_maples = hopkins_trees.where("common name", "Maple, red")
red_maples.sort("count", descending=True)
| plot | common name | count |
|---|---|---|
| p0621 | Maple, red | 106 |
| p1236 | Maple, red | 81 |
| p0821 | Maple, red | 76 |
| p1032 | Maple, red | 72 |
| p0629 | Maple, red | 65 |
| p1133 | Maple, red | 64 |
| p1141 | Maple, red | 64 |
| p0630 | Maple, red | 63 |
| p0622 | Maple, red | 62 |
| p0940 | Maple, red | 61 |
... (341 rows omitted)
But where are those plots?#
plot_info = Table.read_table("data/hopkins-plots.csv").select("plot", "latitude", "longitude")
plot_info
| plot | latitude | longitude |
|---|---|---|
| p00-1 | 42.7472 | -73.2759 |
| p00-2 | 42.7472 | -73.2772 |
| p0000 | 42.7472 | -73.2747 |
| p0001 | 42.7472 | -73.2735 |
| p0002 | 42.7472 | -73.2723 |
| p0003 | 42.7472 | -73.271 |
| p0004 | 42.7472 | -73.2698 |
| p0005 | 42.7472 | -73.2686 |
| p0006 | 42.7472 | -73.2673 |
| p0007 | 42.7472 | -73.2661 |
... (413 rows omitted)
red_maples.join("plot", plot_info)
| plot | common name | count | latitude | longitude |
|---|---|---|---|---|
| p00-1 | Maple, red | 8 | 42.7472 | -73.2759 |
| p00-2 | Maple, red | 2 | 42.7472 | -73.2772 |
| p0000 | Maple, red | 13 | 42.7472 | -73.2747 |
| p0001 | Maple, red | 20 | 42.7472 | -73.2735 |
| p0002 | Maple, red | 12 | 42.7472 | -73.2723 |
| p0003 | Maple, red | 4 | 42.7472 | -73.271 |
| p0004 | Maple, red | 2 | 42.7472 | -73.2698 |
| p0005 | Maple, red | 5 | 42.7472 | -73.2686 |
| p0006 | Maple, red | 3 | 42.7472 | -73.2673 |
| p0007 | Maple, red | 6 | 42.7472 | -73.2661 |
... (341 rows omitted)
Visualization!#
trees_with_lat_lon = hopkins_trees.join("plot", plot_info)
def population_map(tree_name):
counts = trees_with_lat_lon.where("common name", tree_name)
counts = counts.select("latitude", "longitude", "count")
points = counts.with_columns("colors", "blue",
"areas", 1.0 * counts.column("count")).drop("count")
return Circle.map_table(points)
population_map("Maple, red")
Make this Notebook Trusted to load map: File -> Trust Notebook
Exploration, Hypotheses, and Drawing Conclusions#
all_tree_names = np.unique(np.sort(trees_with_lat_lon.column("common name")))
interact(population_map, tree_name=Choice(all_tree_names))
Even More Visualization#
def population_choropleth(tree_name):
counts = trees_with_lat_lon.where("common name", tree_name).select("plot", "count")
return HopkinsForest.map_table(counts)
population_choropleth("Maple, red")
Make this Notebook Trusted to load map: File -> Trust Notebook

