Linear Regression¶
from datascience import *
%matplotlib inline
import matplotlib.pyplot as plots
plots.style.use('fivethirtyeight')
import numpy as np
import warnings
warnings.simplefilter(action='ignore', category=np.VisibleDeprecationWarning)
warnings.simplefilter(action='ignore', category=UserWarning)
from ipywidgets import interact, interactive, fixed, interact_manual, FloatSlider
import ipywidgets as widgets
from mpl_toolkits import mplot3d
import matplotlib.colors as colors
Note!¶
This lecture develops our linear regression code in stages. If you’d like to use linear regression in your labs or projects, you can skip the the last section, which contains all of the linear regression code in one cell.
1. Lines on scatter plots¶
#load data from the lecture 7 and clean it up
greenland_climate = Table().read_table('data/climate_upernavik.csv')
greenland_climate = greenland_climate.relabeled('Precipitation (millimeters)', "Precip (mm)")
tidy_greenland = greenland_climate.where('Air temperature (C)', are.not_equal_to(999.99))
tidy_greenland = tidy_greenland.where('Sea level pressure (mbar)', are.not_equal_to(9999.9))
tidy_greenland.show(3)
| Year | Month | Air temperature (C) | Sea level pressure (mbar) | Precip (mm) |
|---|---|---|---|---|
| 1874 | 9 | 0.1 | 1010.7 | 68 |
| 1874 | 10 | -5.4 | 1002.7 | 24 |
| 1874 | 11 | -8 | 1010.5 | 15 |
... (1256 rows omitted)
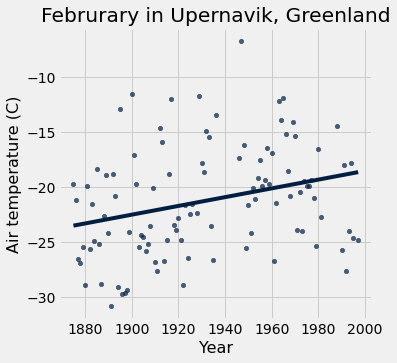
feb = tidy_greenland.where('Month', are.equal_to(2))
feb.scatter('Year', 'Air temperature (C)', fit_line=True)
plots.title('Februrary in Upernavik, Greenland');
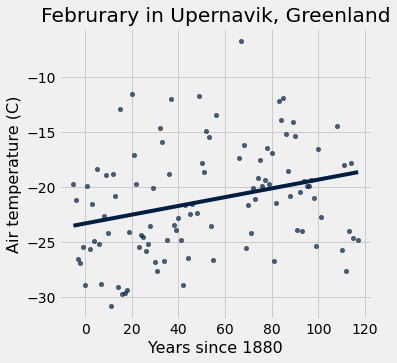
#redo table w/ x-axis as Years since 1880
x = feb.column('Year') - 1880
y = feb.column('Air temperature (C)')
x_y_table = Table().with_columns("x", x, "y", y)
x_y_table.scatter("x", "y", fit_line=True)
plots.xlabel("Years since 1880")
plots.ylabel("Air temperature (C)");
plots.title('Februrary in Upernavik, Greenland');
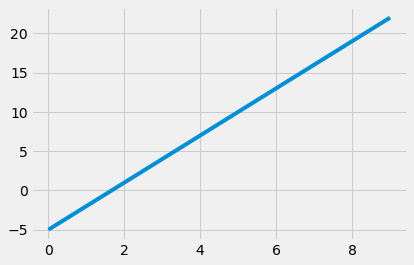
def plot_line(a, b, x):
"""
Plots a line using the give slope and intercept over the
range of x-values in the x array.
"""
xlims = make_array(min(x), max(x))
plots.plot(xlims, a * xlims + b, lw=4)
plot_line(3, -5, np.arange(0, 10))
# Manually put in a, b
rise = -19+24
run = 120
a = rise/run
b = -24
print("a=", a, " , b=", b)
a= 0.041666666666666664 , b= -24
# Plot our manual a and b
x_y_table.scatter("x", "y")
plot_line(a, b, x_y_table.column("x"))
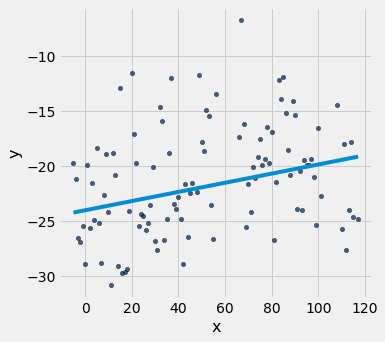
# Pick different lines:
def scatter_with_line(table, x_label, y_label, a, b):
table.scatter(x_label, y_label)
plots.ylim(min(table.column(y_label)), max(table.column(y_label)))
plot_line(a, b, table.column(x_label))
plots.title('a = ' + str(round(a,3)) + '; b = ' + str(round(b,3)));
scatter_with_line(x_y_table, 'x', 'y', a, b)
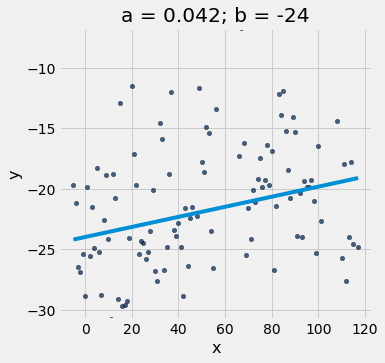
A different slope and intercept
scatter_with_line(x_y_table, 'x', 'y', 0.06, -20)
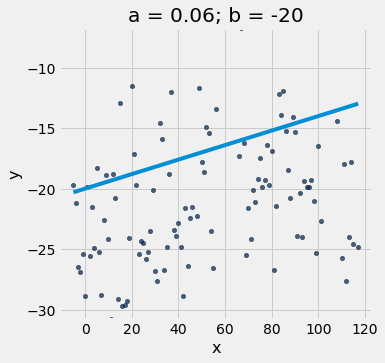
If you run the notebook in Jupyter, you can interact with our interactive visualizations…
def visualize_a_b():
_ = widgets.interact(scatter_with_line,
table = fixed(x_y_table),
x_label = fixed('x'),
y_label = fixed('y'),
a = (-1,1,0.01),
b = (-35,-8,0.1))
visualize_a_b()
2. Predict and evaluate¶
def line_predictions(a, b, x):
"""
Computes the prediction y_hat = a * x + b
where a and b are the slope and intercept and x is the set of x-values.
"""
return a*x + b
y_hat = line_predictions(a, b, x_y_table.column("x"))
prediction_table = x_y_table.with_columns("y_hat", y_hat)
prediction_table.take(np.arange(10, 15))
| x | y | y_hat |
|---|---|---|
| 5 | -18.3 | -23.7917 |
| 6 | -25.2 | -23.75 |
| 7 | -28.8 | -23.7083 |
| 8 | -22.6 | -23.6667 |
| 9 | -18.9 | -23.625 |
# create residuals
residual = prediction_table.column("y") - prediction_table.column("y_hat")
prediction_table = prediction_table.with_columns('residual', residual)
prediction_table.show(3)
| x | y | y_hat | residual |
|---|---|---|---|
| -5 | -19.7 | -24.2083 | 4.50833 |
| -4 | -21.2 | -24.1667 | 2.96667 |
| -3 | -26.5 | -24.125 | -2.375 |
... (102 rows omitted)
def scatter_with_line_and_residuals(table, x_label, y_label, a, b):
"""
Draw a scatter plot, a line, and the line segments capturing the residuals
for some of the points.
"""
y_hat = line_predictions(a, b, table.column(x_label))
prediction_table = x_y_table.with_columns(y_label + '_hat', y_hat)
scatter_with_line(table, x_label, y_label, a, b)
every10th = prediction_table.sort(x_label).take(np.arange(0,
prediction_table.num_rows,
prediction_table.num_rows // 10))
for row in every10th.rows:
x = row.item(x_label)
y = row.item(y_label)
plots.plot([x, x], [y, a * x + b], color='r', lw=2)
scatter_with_line_and_residuals(x_y_table, 'x', 'y', 0.06, -20)
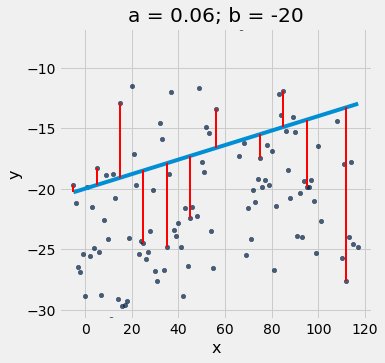
def visualize_residuals():
_ = widgets.interact(scatter_with_line_and_residuals,
table = fixed(x_y_table),
x_label = fixed('x'),
y_label = fixed('y'),
a = (-1,1,0.01),
b = (-35,-8,0.1))
visualize_residuals()
# Mean squared error
np.mean(residual**2)
22.57361111111111
def mean_squared_error(table, x_label, y_label, a, b):
y_hat = line_predictions(a, b, table.column(x_label))
residual = table.column(y_label) - y_hat
return np.mean(residual**2)
mean_squared_error(x_y_table, 'x', 'y', a, b)
22.57361111111111
mean_squared_error(x_y_table, 'x', 'y', 0.04, -24)
22.683558095238094
mean_squared_error(x_y_table, 'x', 'y', 0.02, -20)
27.799756190476195
def scatter_with_line_and_mse(table, x_label, y_label, a, b):
scatter_with_line_and_residuals(table, x_label, y_label, a, b)
mse = mean_squared_error(x_y_table, x_label, y_label, a, b)
plots.title('a = ' + str(round(a,3)) +
'; b = ' + str(round(b,3)) +
'\n mse = ' + str(round(mse,3)));
def visualize_mse():
_ = widgets.interact(scatter_with_line_and_mse,
table = fixed(x_y_table),
x_label = fixed('x'),
y_label = fixed('y'),
a = (-1,1,0.01),
b = (-35,-8,0.1))
visualize_mse()
3. Fit the Best Line Manually¶
a_space = np.linspace(-0.5, 0.5, 200)
b_space = np.linspace(-35, -8, 200)
a_space, b_space = np.meshgrid(a_space, b_space)
broadcasted = np.broadcast(a_space, b_space)
mses = np.empty(broadcasted.shape)
mses.flat = [mean_squared_error(x_y_table, 'x', 'y', a, b) for (a,b) in broadcasted]
def scatter_with_line_and_mse_heat(table, x_label, y_label, a, b, show_mse=None):
plots.figure(figsize=(12,6))
plots.subplot(1, 2, 1)
plots.scatter(table.column(x_label), table.column(y_label),color='darkslateblue',s=30)
plots.ylim(min(table.column(y_label)), max(table.column(y_label)))
plots.xlabel('x')
plots.ylabel('y')
plot_line(a, b, table.column(x_label))
mse = mean_squared_error(table, x_label, y_label, a, b)
plots.title('a = ' + str(round(a,3)) +
'; b = ' + str(round(b,3)) +
'\nmse = ' + format(mse, '.3f'))
if show_mse == '2d':
plots.subplot(1, 2, 2)
plots.pcolormesh(a_space, b_space, mses,
norm=colors.PowerNorm(gamma=0.25,
vmin=mses.min(),
vmax=mses.max()));
plots.scatter(a,b,s=100,color='red');
plots.xlabel('a')
plots.ylabel('b')
elif show_mse == '3d':
ax = plots.subplot(1, 2, 2, projection='3d')
ax.plot_surface(a_space, b_space, mses,
cmap='viridis',
antialiased=False,
linewidth=0,
norm = colors.PowerNorm(gamma=0.25,vmin=mses.min(), vmax=mses.max()))
ax.plot([a],[b],[mean_squared_error(x_y_table, 'x', 'y', a, b)], 'ro');
ax.set_xlabel('a')
ax.set_ylabel('b')
plots.tight_layout()
def visualize_full_mse():
_ = widgets.interact(scatter_with_line_and_mse_heat,
table = fixed(x_y_table),
x_label = fixed('x'),
y_label = fixed('y'),
a = (-0.5,0.5,0.01),
b = (-35,-8,0.1),
show_mse=[None, "2d", "3d"])
visualize_full_mse()
4. Fit the Best Line with Brute Force¶
# Brute force loop to find a and b
lowest_mse = np.inf
a_for_lowest = 0
b_for_lowest = 0
for a in np.arange(0.02, 0.08, 0.001):
for b in np.arange(-30, 30, 1):
mse = mean_squared_error(x_y_table, 'x', 'y', a, b)
if mse < lowest_mse:
lowest_mse = mse
a_for_lowest = a
b_for_lowest = b
lowest_mse, a_for_lowest, b_for_lowest
(22.216645485714285, 0.03600000000000002, -23)
# Looks pretty good when we plot it!
scatter_with_line_and_mse_heat(x_y_table, 'x', 'y', a_for_lowest, b_for_lowest, show_mse="2d")
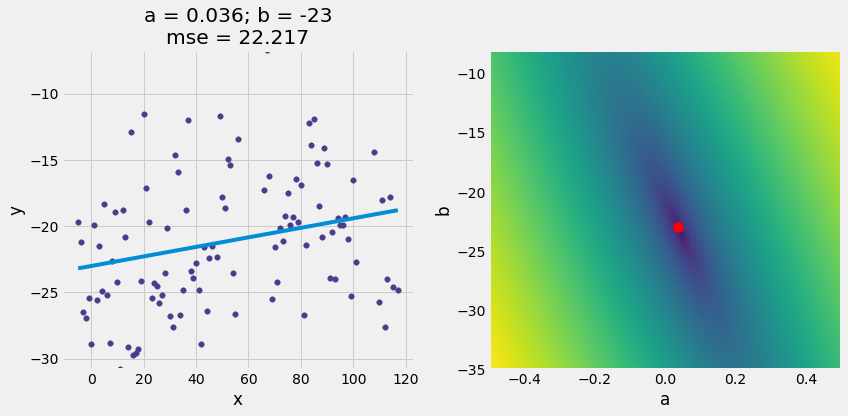
5. Fit the Best Line with minimize()¶
def linear_regression(table, x_label, y_label):
"""
Return an array containing the slope and intercept of the line best fitting
the table's data according to the mean square error loss function.
"""
# A helper function that takes *only* the two variables we need to optimize.
# This is necessary to use minimize below, because the function we want
# to minimize cannot take any parameters beyond those it will solve for.
def mse_for_a_b(a, b):
return mean_squared_error(table, x_label, y_label, a, b)
# We can use a built-in function `minimize` that uses gradiant descent
# to find the optimal a and b for our mean squared error function
return minimize(mse_for_a_b)
optimal = linear_regression(x_y_table, 'x', 'y')
a_optimal = optimal.item(0)
b_optimal = optimal.item(1)
scatter_with_line_and_mse_heat(x_y_table, 'x', 'y', a_optimal, b_optimal, show_mse="2d")
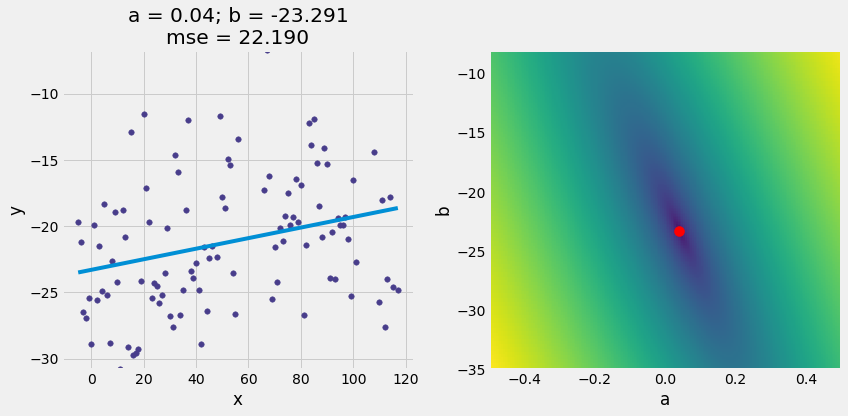
6. Diagnostics¶
def calculate_residuals(x, y, a, b):
y_hat = line_predictions(a, b, x)
residual = y - y_hat
return residual
def r2_score(table, x_label, y_label, a, b):
x = table.column(x_label)
y = table.column(y_label)
y_mean = np.mean(y)
residual = calculate_residuals(x, y, a, b)
numerator = sum(residual**2)
denominator = sum((y-y_mean)**2)
return 1 - numerator/denominator
r2_score(x_y_table, 'x', 'y', a_optimal, b_optimal)
0.08285898226812227
Residual plots¶
Residual plots help us make visual assessments of the quality of a linear regression analysis
def residual_plot(table, x_label, y_label, a, b):
x = table.column(x_label)
y = table.column(y_label)
residual = calculate_residuals(x, y, a, b)
t = Table().with_columns('x', x, 'residuals', residual)
t.scatter('x', 'residuals', color='r')
xlims = make_array(min(x), max(x))
plots.plot(xlims, make_array(0, 0), color='darkblue', lw=4)
plots.title('Residual Plot')
residual_plot(x_y_table, 'x', 'y', a_optimal, b_optimal)
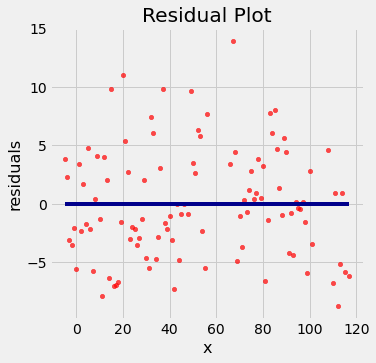
Notice how the residuals are distributed fairly symmetrically above and below the horizontal line at 0, corresponding to the original scatter plot being roughly symmetrical above and below.
The residual plot of a good regression shows no pattern. The residuals look about the same, above and below the horizontal line at 0, across the range of the predictor variable.
Diagnostics and residual plot on other datasets
cars = Table().read_table('data/hybrid.csv')
cars.show(3)
| vehicle | year | msrp | acceleration | mpg | class |
|---|---|---|---|---|---|
| Prius (1st Gen) | 1997 | 24509.7 | 7.46 | 41.26 | Compact |
| Tino | 2000 | 35355 | 8.2 | 54.1 | Compact |
| Prius (2nd Gen) | 2000 | 26832.2 | 7.97 | 45.23 | Compact |
... (150 rows omitted)
# Fit the line to the data
a_cars, b_cars = linear_regression(cars, 'acceleration', 'mpg')
# Plot the regression line
scatter_with_line(cars, 'acceleration', 'mpg', a_cars, b_cars)
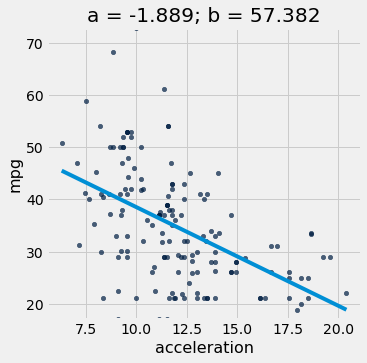
r2_score(cars, 'acceleration', 'mpg', a_cars, b_cars)
0.25610723394360513
#plot the residuals
residual_plot(cars, 'acceleration', 'mpg', a_cars, b_cars)
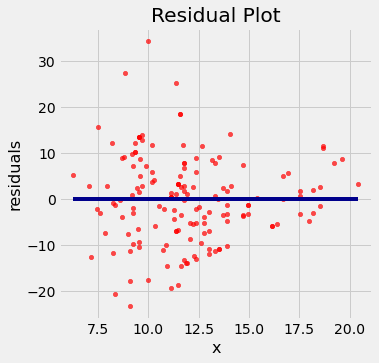
If the residual plot shows uneven variation about the horizontal line at 0, the regression estimates are not equally accurate across the range of the predictor variable.
The residual plot shows that our linear regression for this dataset is heteroscedastic, that x and the residuals are not independent.
7. The Complete Code¶
#
# The complete CS 104 linear regresion code!
#
def plot_line(a, b, x):
"""
Plots a line using the give slope and intercept over the
range of x-values in the x array.
"""
xlims = make_array(min(x), max(x))
plots.plot(xlims, a * xlims + b, lw=4)
def scatter_with_line(table, x_label, y_label, a, b):
"""
Draw a scatter plot of the given table data overlaid with
a line having the given slope a and intercept b.
"""
table.scatter(x_label, y_label)
plot_line(a, b, table.column(x_label))
plots.title('a = ' + str(round(a,3)) + '; b = ' + str(round(b,3)));
def line_predictions(a, b, x):
"""
Computes the prediction y_hat = a * x + b
where a and b are the slope and intercept and x is the set of x-values.
"""
return a * x + b
def mean_squared_error(table, x_label, y_label, a, b):
"""
Returns the mean squared error for the line described by slope a and
intercept b when used to fit the data in the tables x_label and y_label
columns.
"""
y_hat = line_predictions(a, b, table.column(x_label))
residual = table.column(y_label) - y_hat
return np.mean(residual**2)
def linear_regression(table, x_label, y_label):
"""
Return an array containing the slope and intercept of the line best fitting
the table's data according to the mean square error loss function.
"""
# A helper function that takes *only* the two variables we need to optimize.
# This is necessary to use minimize below, because the function we want
# to minimize cannot take any parameters beyond those it will solve for.
def mse_for_a_b(a, b):
return mean_squared_error(table, x_label, y_label, a, b)
# We can use a built-in function `minimize` that uses gradiant descent
# to find the optimal a and b for our mean squared error function
return minimize(mse_for_a_b)
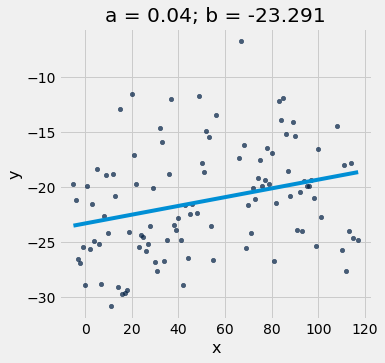
# Example using the linear regression code
optimal = linear_regression(x_y_table, 'x', 'y')
a_optimal = optimal.item(0)
b_optimal = optimal.item(1)
print(a_optimal, b_optimal)
scatter_with_line(x_y_table, 'x', 'y', a_optimal, b_optimal)
0.03988600486015187 -23.291236810681426
Extras for Slides¶
def pearson_correlation(table, x_label, y_label):
x = table.column(x_label)
y = table.column(y_label)
x_mean = np.mean(x)
y_mean = np.mean(y)
numerator = sum((x - x_mean) * (y - y_mean))
denominator = np.sqrt(sum((x - x_mean)**2)) * np.sqrt(sum((y - y_mean)**2))
return numerator / denominator
def plot_scatter_along_line(a, b, num_points=100):
xs = np.random.uniform(0,10,100)
ys = [ a*x + b + np.random.normal(0,0.5) for x in xs]
t = Table().with_columns('x', xs, 'y', ys)
print(pearson_correlation(t, 'x', 'y'))
plots.scatter(xs,ys)
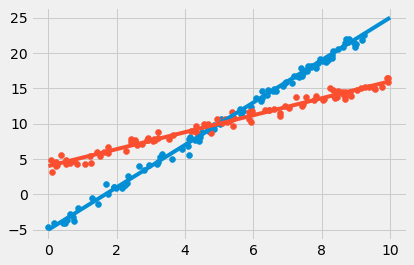
plot_scatter_along_line(3,-5)
plot_scatter_along_line(1.2, 4)
plot_line(3, -5, np.arange(0, 10.1))
plot_line(1.2, 4, np.arange(0, 10.1))
0.9981673683123793
0.9897478621282604