Causality
Contents
Causality#
from datascience import *
from cs104 import *
import numpy as np
%matplotlib inline
1. Load maternal smoker data#
You can read more about this data here https://www.stat.berkeley.edu/~statlabs/labs.html#babiesI
births = Table.read_table('data/baby.csv')
births
| Birth Weight | Gestational Days | Maternal Age | Maternal Height | Maternal Pregnancy Weight | Maternal Smoker |
|---|---|---|---|---|---|
| 120 | 284 | 27 | 62 | 100 | False |
| 113 | 282 | 33 | 64 | 135 | False |
| 128 | 279 | 28 | 64 | 115 | True |
| 108 | 282 | 23 | 67 | 125 | True |
| 136 | 286 | 25 | 62 | 93 | False |
| 138 | 244 | 33 | 62 | 178 | False |
| 132 | 245 | 23 | 65 | 140 | False |
| 120 | 289 | 25 | 62 | 125 | False |
| 143 | 299 | 30 | 66 | 136 | True |
| 140 | 351 | 27 | 68 | 120 | False |
... (1164 rows omitted)
births.show(4)
| Birth Weight | Gestational Days | Maternal Age | Maternal Height | Maternal Pregnancy Weight | Maternal Smoker |
|---|---|---|---|---|---|
| 120 | 284 | 27 | 62 | 100 | False |
| 113 | 282 | 33 | 64 | 135 | False |
| 128 | 279 | 28 | 64 | 115 | True |
| 108 | 282 | 23 | 67 | 125 | True |
... (1170 rows omitted)
smoking_and_birthweight = births.select('Maternal Smoker', 'Birth Weight')
smoking_and_birthweight.group('Maternal Smoker')
| Maternal Smoker | count |
|---|---|
| False | 715 |
| True | 459 |
smoking_and_birthweight.hist('Birth Weight', group='Maternal Smoker')
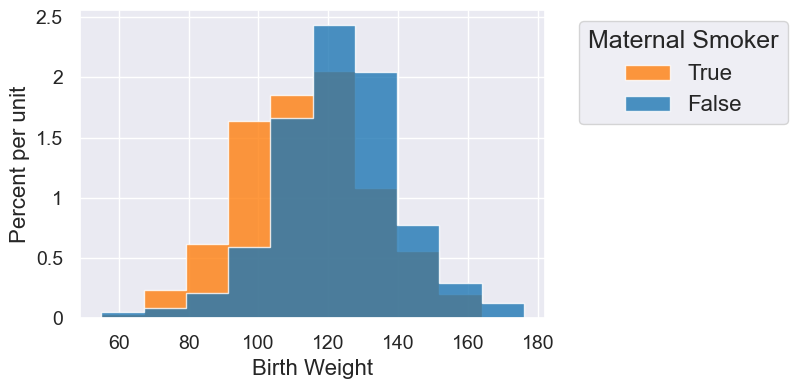
2. Test Statistic#
means_table = smoking_and_birthweight.group('Maternal Smoker', np.mean)
means_table
| Maternal Smoker | Birth Weight mean |
|---|---|
| False | 123.085 |
| True | 113.819 |
means = means_table.column('Birth Weight mean')
observed_difference = means.item(1) - means.item(0)
observed_difference
-9.266142572024918
In keeping with the approach we laid out last lecture, we’ll focus only on absolute difference…
observed_difference = abs(means.item(1) - means.item(0))
observed_difference
9.266142572024918
def abs_difference_of_means(table, group_label, value_label):
"""
Inputs:
- table: name of table
- group_label: column label for variable you want to group
- value_label: column label for the variable you want to take means of
Returns: Difference of mean birth weights of the two groups
"""
# table containing group means
means_table = table.group(group_label, np.mean)
# array of group means
means = means_table.column(value_label + ' mean')
return abs(means.item(1) - means.item(0))
Our observed difference
observed_difference = abs_difference_of_means(births, 'Maternal Smoker', "Birth Weight")
observed_difference
9.266142572024918
We can use this function on lots of columns!
abs_difference_of_means(births, 'Maternal Smoker', "Maternal Age")
0.8076725017901509
abs_difference_of_means(births, 'Maternal Smoker', "Maternal Height")
0.09058914941267915
3. Simulation Under Null Hypothesis#
Creating Permutations of Labels#
Just use a tiny table to show our approach…
tiny_smoking_and_birthweight = smoking_and_birthweight.take(np.arange(0,6))
shuffled_labels = tiny_smoking_and_birthweight.sample(with_replacement=False).column('Maternal Smoker')
original_and_shuffled = tiny_smoking_and_birthweight.with_column('Shuffled Label', shuffled_labels)
original_and_shuffled
| Maternal Smoker | Birth Weight | Shuffled Label |
|---|---|---|
| False | 120 | False |
| False | 113 | True |
| True | 128 | True |
| True | 108 | False |
| False | 136 | False |
| False | 138 | False |
A function to make a permutation!
def permutation_sample(table, group_column_name):
"""
Returns: The table with a new "Shuffled Label" column containing
the shuffled values of the group column.
"""
# array of shuffled labels
shuffled_labels = table.sample(with_replacement=False).column(group_column_name)
# table of numerical variable and shuffled labels
shuffled_table = table.with_column('Shuffled Label', shuffled_labels)
return shuffled_table
original_and_shuffled = permutation_sample(tiny_smoking_and_birthweight, "Maternal Smoker")
original_and_shuffled
| Maternal Smoker | Birth Weight | Shuffled Label |
|---|---|---|
| False | 120 | True |
| False | 113 | False |
| True | 128 | False |
| True | 108 | True |
| False | 136 | False |
| False | 138 | False |
# Steve: The statistic for our reshuffled label
abs_difference_of_means(original_and_shuffled, "Shuffled Label", "Birth Weight")
14.75
And now the full table…
smoking_and_birthweight
| Maternal Smoker | Birth Weight |
|---|---|
| False | 120 |
| False | 113 |
| True | 128 |
| True | 108 |
| False | 136 |
| False | 138 |
| False | 132 |
| False | 120 |
| True | 143 |
| False | 140 |
... (1164 rows omitted)
original_and_shuffled = permutation_sample(smoking_and_birthweight, "Maternal Smoker")
original_and_shuffled
| Maternal Smoker | Birth Weight | Shuffled Label |
|---|---|---|
| False | 120 | False |
| False | 113 | False |
| True | 128 | True |
| True | 108 | False |
| False | 136 | False |
| False | 138 | False |
| False | 132 | False |
| False | 120 | True |
| True | 143 | False |
| False | 140 | False |
... (1164 rows omitted)
# "one trial" of our simulation
abs_difference_of_means(original_and_shuffled, 'Shuffled Label', 'Birth Weight')
0.08691743985860967
Permutation Test#
# Steve: I removed the values column name from here -- no need to include it and do a select
# Just return the original table with a new column.
def permutation_sample(table, group_column_name):
"""
Returns: The table with a new "Shuffled Label" column containing
the shuffled values of the group column.
"""
# array of shuffled labels
shuffled_labels = table.sample(with_replacement=False).column(group_column_name)
# table of numerical variable and shuffled labels
shuffled_table = table.with_column('Shuffled Label', shuffled_labels)
return shuffled_table
def simulate_birth_weights(num_trials):
birth_weight_simulated_statistics = make_array()
for i in np.arange(0, num_trials):
one_sample = permutation_sample(smoking_and_birthweight, "Maternal Smoker")
statistic_one_sample = abs(abs_difference_of_means(one_sample,
"Shuffled Label",
"Birth Weight"))
birth_weight_simulated_statistics = np.append(birth_weight_simulated_statistics,
statistic_one_sample)
return birth_weight_simulated_statistics
simulated_birth_weight_diffs = simulate_birth_weights(1000)
results = Table().with_column('abs(Group B Mean - Group A Mean)', simulated_birth_weight_diffs)
results.hist().dot(observed_difference)
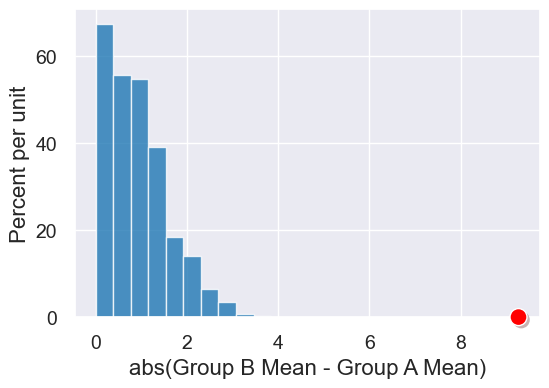
Let’s calculate the p-value (even if we can easily guess what it is here)…
sum(simulated_birth_weight_diffs >= observed_difference) / len(simulated_birth_weight_diffs)
0.0
4. Randomized Controlled Experiment#
rct = Table.read_table('data/bta.csv')
rct.show()
| Group | Result |
|---|---|
| Control | 1 |
| Control | 1 |
| Control | 0 |
| Control | 0 |
| Control | 0 |
| Control | 0 |
| Control | 0 |
| Control | 0 |
| Control | 0 |
| Control | 0 |
| Control | 0 |
| Control | 0 |
| Control | 0 |
| Control | 0 |
| Control | 0 |
| Control | 0 |
| Treatment | 1 |
| Treatment | 1 |
| Treatment | 1 |
| Treatment | 1 |
| Treatment | 1 |
| Treatment | 1 |
| Treatment | 1 |
| Treatment | 1 |
| Treatment | 1 |
| Treatment | 0 |
| Treatment | 0 |
| Treatment | 0 |
| Treatment | 0 |
| Treatment | 0 |
| Treatment | 0 |
rct.pivot('Result', 'Group') #Recall: the default aggregation function counts the items
| Group | 0.0 | 1.0 |
|---|---|---|
| Control | 14 | 2 |
| Treatment | 6 | 9 |
# This tells us the proportion in each group who improved.
rct.group('Group', np.average)
| Group | Result average |
|---|---|
| Control | 0.125 |
| Treatment | 0.6 |
Testing the Hypotheses#
shuffled_labels = rct.sample(with_replacement=False).column('Group')
original_and_shuffled = rct.with_column('Shuffled Label', shuffled_labels)
original_and_shuffled
| Group | Result | Shuffled Label |
|---|---|---|
| Control | 1 | Control |
| Control | 1 | Control |
| Control | 0 | Treatment |
| Control | 0 | Treatment |
| Control | 0 | Treatment |
| Control | 0 | Treatment |
| Control | 0 | Control |
| Control | 0 | Control |
| Control | 0 | Treatment |
| Control | 0 | Control |
... (21 rows omitted)
# Original RCT
original_and_shuffled.select('Result', 'Group').group('Group', np.average)
| Group | Result average |
|---|---|
| Control | 0.125 |
| Treatment | 0.6 |
# Shuffled RCT from the permutation test
original_and_shuffled.select('Result', 'Shuffled Label').group('Shuffled Label', np.average)
| Shuffled Label | Result average |
|---|---|
| Control | 0.3125 |
| Treatment | 0.4 |
def difference_of_proportions(table, group_label, value_label):
"""Takes: name of table,
column label that indicates which group the row relates to
Returns: Difference of proportions of 1's in the two groups"""
# table containing group means
proportions_table = table.group(group_label, np.mean)
# array of group means
proportions = proportions_table.column(value_label + ' mean')
return abs(proportions.item(1) - proportions.item(0))
observed_diff = difference_of_proportions(rct, 'Group', 'Result')
observed_diff
0.475
# Reuse permutation_sample, and structure same as first example.
def empirical_difference_of_proportions_distribution(num_iterations):
simulated_diffs = make_array()
for i in np.arange(num_iterations):
one_sample = permutation_sample(rct, 'Group')
statistic_one_sample = abs(abs_difference_of_means(one_sample,
"Shuffled Label",
'Result'))
simulated_diffs = np.append(simulated_diffs, statistic_one_sample)
return simulated_diffs
simulated_diffs = empirical_difference_of_proportions_distribution(2000)
col_name = 'Difference between Treatment and Control'
results = Table().with_column(col_name, simulated_diffs)
plot = results.hist(bins=np.arange(0,0.7,0.1))
plot.dot(observed_diff)
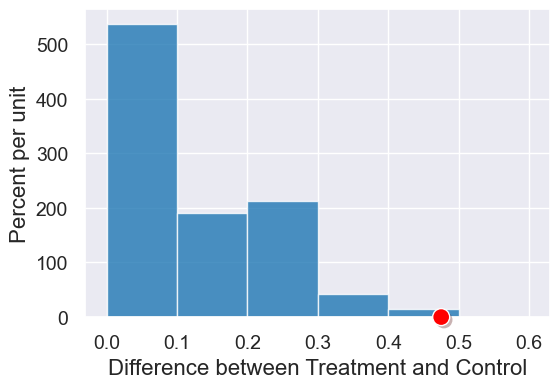
# p-value
sum(simulated_diffs >= observed_diff) / len(simulated_diffs)
0.0065

